Decision Trees and Random Forest
STAT 220
Decision Tree
A decision tree algorithm learns by repeatedly splitting the dataset into increasingly smaller subsets to accurately predict the target value.
Data is continuously split according to a certain parameter
Two main entities:
- nodes: where the data is split
- leaves: decisions or final outcomes
Decision Tree
Use features to make subsets of cases that are as similar (“pure”) as possible with respect to the outcome
- Start with all observations in one group
- Find the variable/feature/split that best separates the outcome
- Divide the data into two groups (leaves) on the split (node)
- Within each split, find the best variable/split that separates the outcomes
- Continue until the groups are too small or sufficiently “pure”
Data preparation and pre-processing
data(PimaIndiansDiabetes2)
db <- PimaIndiansDiabetes2 %>% drop_na() %>%
mutate(diabetes = fct_relevel(diabetes, c("neg", "pos")))
set.seed(314)
db_split <- initial_split(db, prop = 0.75)
db_train <- db_split %>% training()
db_test <- db_split %>% testing()
# scaling not needed
db_recipe <- recipe(diabetes ~ ., data = db_train) %>%
step_dummy(all_nominal(), -all_outcomes()) Model Specification
- cost_complexity: The cost complexity parameter, the minimum improvement in the model needed at each node
- tree_depth: The maximum depth of a tree
- min_n: The minimum number of data points in a node that are required for the node to be split further.
Workflow and Hyperparameter tuning
# Combine the model and recipe into a workflow
tree_workflow <- workflow() %>%
add_model(tree_model) %>%
add_recipe(db_recipe)
# Create folds for cross validation on the training data set
db_folds <- vfold_cv(db_train, v = 5, strata = diabetes)
## Create a grid of hyperparameter values to optimize
tree_grid <- grid_random(cost_complexity(),
tree_depth(),
min_n(),
size = 10)View grid
Tuning Hyperparameters with tune_grid()
# Tune decision tree workflow
set.seed(314)
tree_tuning <- tree_workflow %>%
tune_grid(resamples = db_folds,
grid = tree_grid)
# Select best model based on accuracy
best_tree <- tree_tuning %>%
select_best(metric = 'accuracy')
# View the best tree parameters
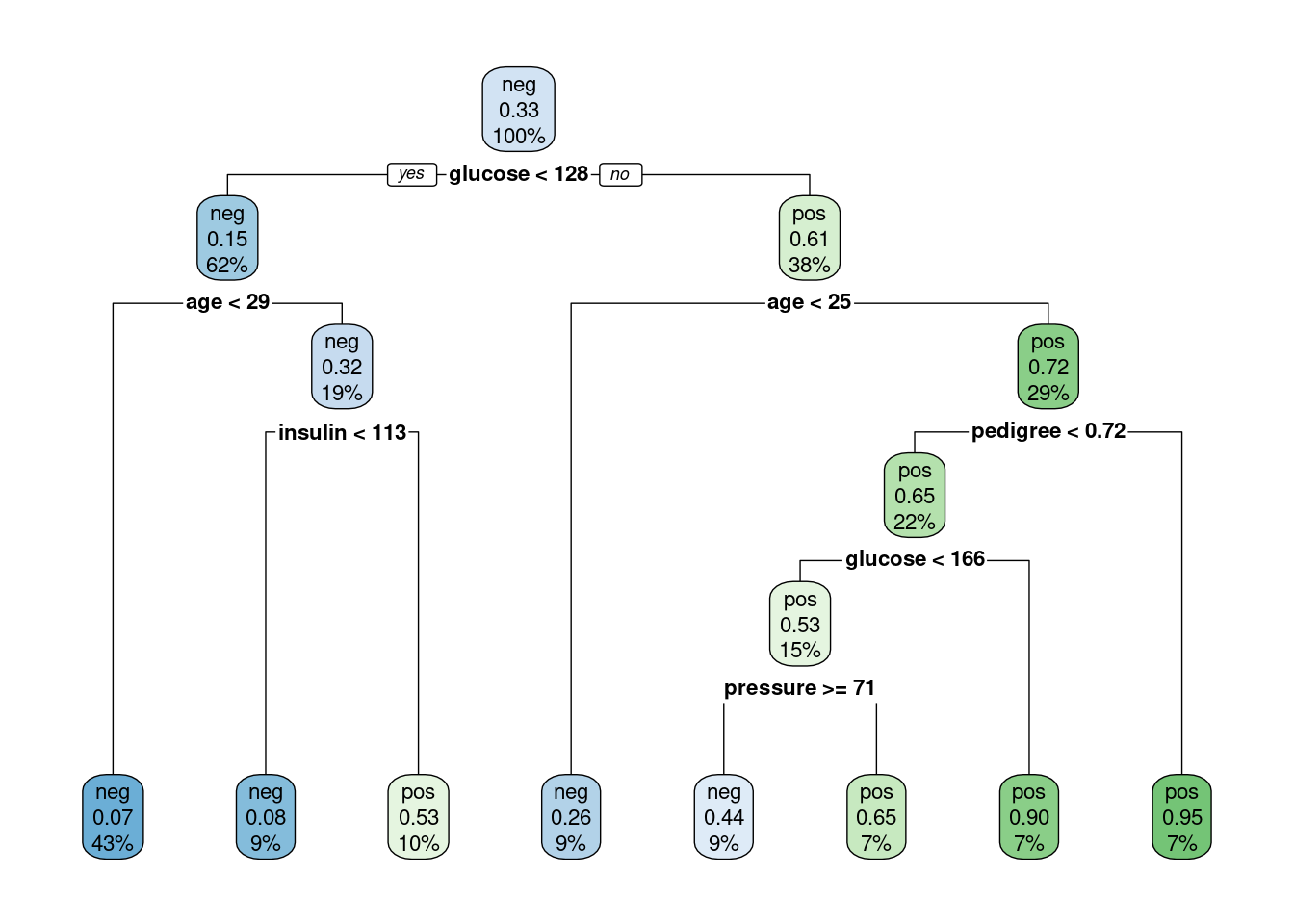
best_tree# A tibble: 1 × 4
cost_complexity tree_depth min_n .config
<dbl> <int> <int> <chr>
1 5.28e-10 7 40 Preprocessor1_Model01Plot the tree
Train and Evaluate With last_fit()
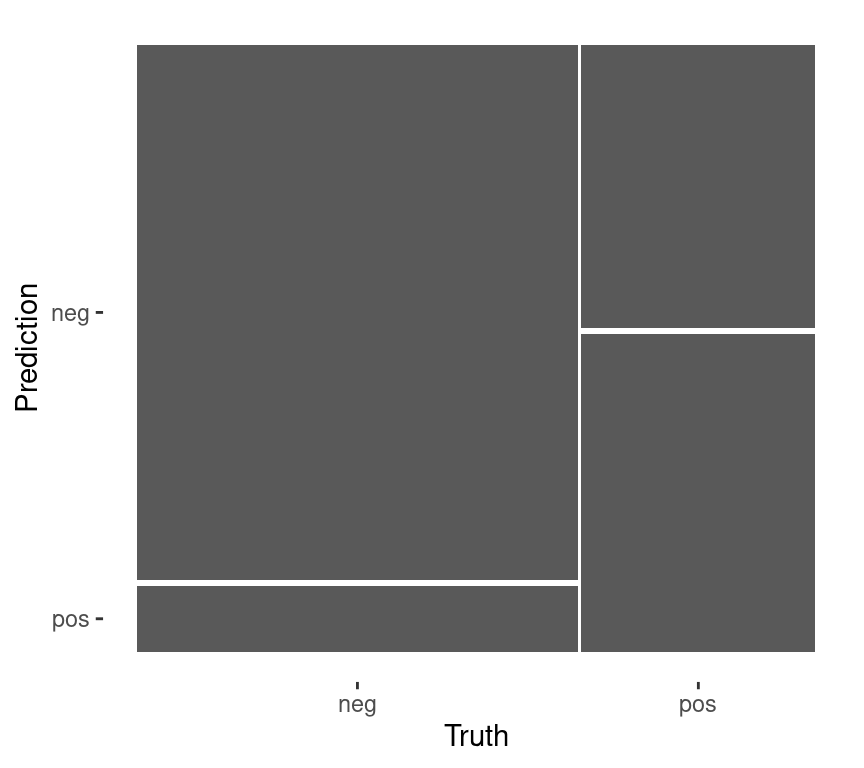
# A tibble: 3 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 accuracy binary 0.765 Preprocessor1_Model1
2 roc_auc binary 0.824 Preprocessor1_Model1
3 brier_class binary 0.153 Preprocessor1_Model1Confusion matrix
Group Activity 1
- Please clone the
ca27-yourusernamerepository from Github - Please do problem 1 in the class activity for today
20:00 Random Forest
Random forests take decision trees and construct more powerful models in terms of prediction accuracy.
-
Repeated sampling (with replacement) of the training data to produce a sequence of decision tree models.
These models are then averaged to obtain a single prediction for a given value in the predictor space.
The random forest model selects a random subset of predictor variables for splitting the predictor space in the tree building process.
Model Specification
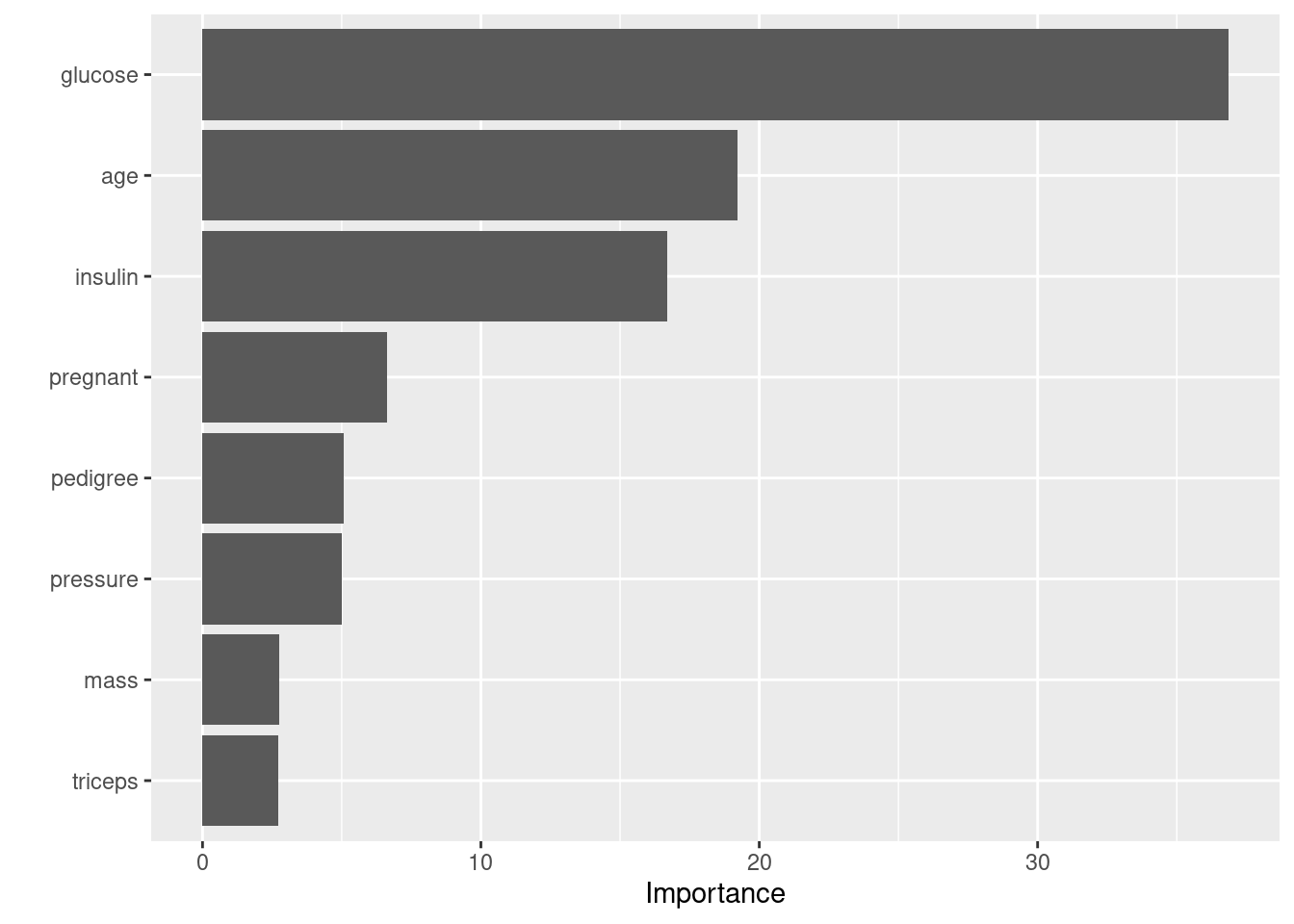
- mtry: The number of predictors that will be randomly sampled at each split when creating the tree models
- trees: The number of decision trees to fit and ultimately average
- min_n: The minimum number of data points in a node that are required for the node to be split further
Model, Workflow and Hyperparameter Tuning
rf_model <- rand_forest(mtry = tune(),
trees = tune(),
min_n = tune()) %>%
set_engine('ranger', importance = "impurity") %>%
set_mode("classification")
rf_workflow <- workflow() %>%
add_model(rf_model) %>%
add_recipe(db_recipe)
## Create a grid of hyperparameter values to test
set.seed(314)
rf_grid <- grid_random(mtry() %>% range_set(c(2, 7)),
trees(),
min_n(),
size = 15)View Grid
Tuning Hyperparameters with tune_grid()
# Tune random forest workflow
set.seed(314)
rf_tuning <- rf_workflow %>%
tune_grid(resamples = db_folds,
grid = rf_grid)
## Select best model based on roc_auc
best_rf <- rf_tuning %>%
select_best(metric = 'accuracy')
# View the best parameters
best_rf# A tibble: 1 × 4
mtry trees min_n .config
<int> <int> <int> <chr>
1 2 1003 24 Preprocessor1_Model15Finalize workflow
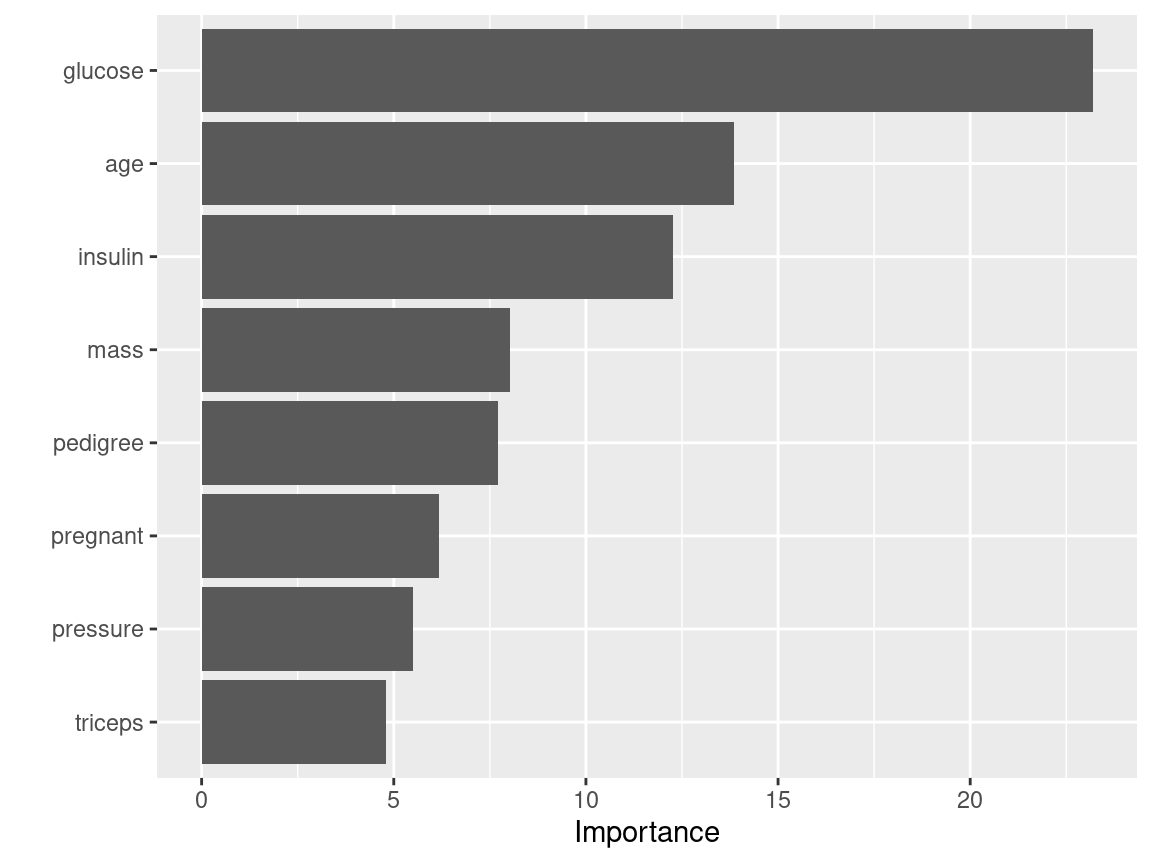
Variable Importance
Group Activity 2

- Please finish the remaining problems in the class activity for today
10:00